The Hep B vax HIV origin theory, pt 1.5
Some... additional... lackluster comments?
*edit, October 3: Commenter TCC alerted me a very helpful recent book on HIV origins, namely Jacques Pépin’s The Origins of AIDS, which led to the identification of some errors in my reconstruction of pre-1980s spread — namely, I had not encountered the overt description of evidence of a lack of spread outside the Congo region before 1980. This post has since been corrected to change references to the virus’s evolution in Africa to “in the Congo region;” I have also corrected the lay description of the type of chimpanzee from which HIV derives.
The following post is intended as a supplemental to Pt. 1, and largely expands on the same points. It intends both to serve my own interest in speculating about genetics and viruses and to offer the reader further avenues to becoming familiar with the topic of HIV in a way that broadly helps ground interpretation of whether it could have come out of medical experiments and vaccines in the 1950s - 1980s. If Pt. 1 was a summary of this same problem, then Pt. 1.5 is intended as largely a series of “put differently”s to flesh out the summary, and will offer extensive quotation of Edward C. Holmes’ remarks on HIV.
As well, this will probably be added to the resource hub.
Again, on the point of cross-over
To recap the argument in Pt. 1, there turned out to be too much diversity in “HIV,” once a larger amount of sequences had been gathered following initial designation of the virus. This was both in the sense of there being multiple groups of HIV and of the predominant group, HIV-1M, having multiple subgroups with deep genetic divergences that must have emerged over time1 — therefore, HIV was already endemic in humans decades before its discovery.
This diversity in 1M did not emerge from the global HIV footprint in the 80s and 90s — it is not, like SARS-CoV-2’s variants, a feature of dispersed mutation acting on a previously singular template while being observed — but simply was revealed to exist in and gradually spilled further out from the Congo-region human HIV reservoir as time went on. As such it was in fact noted early, but not synthesized into the eventual understanding of HIV as a category of multiple viruses which crossed over into humans from primates (at least) many decades before 1980, for some time.
Holmes’ textbook on RNA virus evolution devotes a subchapter to HIV, and since the book is très cher, I will supply the reader overview-scale quotations:2
7.2.2 The genetic diversity of HIV
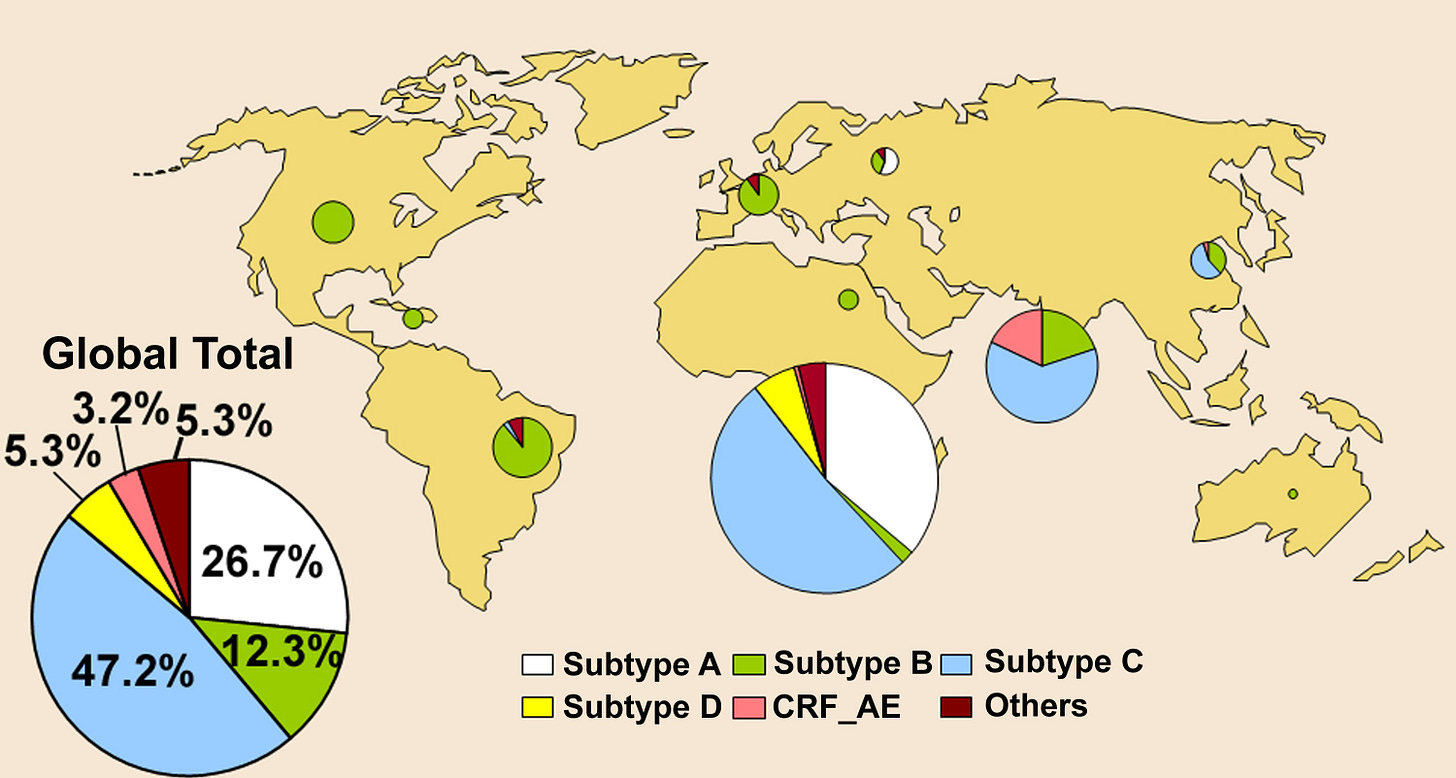
Right from the earliest descriptions of genetic variation in HIV-1 it was clear that this virus was remarkably diverse, both within individual hosts (Hahn et al. 1986; Balfe et al. 1990; Holmes et al. 1992) and globally (Korber et al. 1995). As the worldwide wide sample of HIV-1 began to expand it became clear that viral genetic diversity could often be partitioned into discrete clusters, or clades, on phylogenetic trees, that were eventually christened `subtypes'. At the time of writing there are nine such subtypes types of HIV-1 (denoted A-K, but excluding E and I) […and on to the topic of recombinants, aka CRFs]
The subtypes (and CRFs) of HIV-1 are also notable for their differing geographical distributions: subtype B represents the form of the virus first observed in industrialized nations during the early 1980s and which still dominates in these regions to this day, whereas subtypes A, C, and D are more commonly found in sub-Saharan Africa, with subtype C rising dramatically in frequency, particularly in southern Africa (Fig. 7.7). The other subtypes are found at rather lower frequencies. Phylogenetically defined subtypes of viruses have also been identified in HIV-2, denoted `epidemic' subtypes A and B and non-epidemic subtypes C-G, although all are usually restricted to West Africa (Lemey et al. 2003).
The identification of lentiviruses in a wide range of non-human primates, particularly chimpanzees (Huet et at. 1990; Santiago et al. 2002, 2003; Nerrienet et al. 2005; Keele et al. 2006), changed the evolutionary context of genetic variation in HIV-1. Specifically, the subtypes and CRFs of HIV-1[M] described above fall into a single branch on the HIV phylogeny, reflecting a single cross-species transmission from chimpanzees (Fig. 7.8). This cluster of viruses is denoted the M, or Main, group, and contains the vast majority of viruses assigned to HIV-1. Strikingly, two other clusters (groups) of HIV-1 isolates have been identified, although present at far lower frequencies: 0, for Outlier, and N for New. That these groups are separated from the M group by SIVcpz from chimpanzees [when organized by magic-computer-programs] is powerful evidence not only that chimpanzees are the ultimate source for HIV-1, but that species jumps have occurred a number of times (Fig. 7.8; see below). Also of note is that the greatest phylogenetic diversity in HIV (i.e. HIV-2, the M, N, and 0 groups of HIV-1, and extensive diversity within the M group) is observed in central-west Africa, the most likely birth place for this virus.
I will offer here a preemptive clarification on what is not being said. There are indeed a whole handful of families of HIV, which must each have been introduced into humans from different primate SIV virus crossovers, but HIV-1M is only one of these families, with only one likely cross-over from an ancestral central-African-chimpanzee-SIV3. And HIV-1M is the behemoth of the post-1980s spread of the virus both in Africa and elsewhere. The other families of HIV-1 and 2 essentially do not matter, and so neither really does their origin, except to compare the divergent origins of these families with what is found for subgroups within 1M (that, as said, they all cluster together in a single branch). This will be further discussed below with a reproduction of Fig. 7.8.
Holmes continues:4
7.2.4 The origins and spread of HIV
While some aspects of HIV research have made little progress, particularly the development of vaccines, the study of the origin and spread of HIV has proven remarkably successful.
It would be fair to say that we now know where HIV comes from, with a plausible route of entry into human populations, as well as a rough timescale for these events. In the case of HIV-1 this means that the virus mostly likely emerged in the Congo region of central-west Africa during the first decades of the twentieth century and first entered human populations through exposure to contaminated bush meat from chimpanzees (Gao et al. 1999; Keele et al. 2006; Worobey et al. 2008).
A similar picture can be painted for HIV-2, although the place of emergence is likely to be rather further west in Africa, and sooty mangabeys act as the reservoir species (Santiago et al. 2005).
In both cases there also appears to have been multiple cross-species species transmission events from non-human primates to humans [as in of 1M, 1N, and 1O within the HIV-1 supergroup]. As should be apparent from these statements, documenting the diversity of viruses that circulate in a wide variety of non-human primates is critical to understanding standing the origins of HIV.
Although these are routinely referred to as the SIVs, that none seem to cause overt disease in the natural hosts means that the term immunodeficiency is something of a misnomer, so that the primate lentiviruses is a safer phrase. Indeed, SIV in its natural hosts is not associated with a decline in the number of CD4 T cells despite long-term infection and high levels of viraemia, and does not seem to generate an overly strong immune response (Broussard et at. 2001; Silvestri et at. 2003). Primate lentiviruses are also remarkably abundant. At the time of writing at least 40 of these viruses associated with different primate species have been identified, largely within monkeys of the family Cercopithecidae and apes (chimpanzees, gorillas, las, and humans) of the family Hominidae (Hahn et al. 2000; Santiago et al. 2002; Keele et al. 2006; Van Heuverswyn et al. 2006).
Crucially, these viruses are only found naturally in animals of African origin. Despite the evidence for the antiquity of the lentiviruses as a whole, this phylogenetic distribution strongly suggests that the current lineages of primate lentiviruses were acquired subsequent to the divergence between Old World and New World primates, and that there has been clear species jumping between those viruses that infect Old World monkeys and those that infect [the apes,] chimpanzees, gorillas, and humans (Fig. 7.8).
The fingerprint of a long duration of rarity in 1M
Now we return to 1M, the main group involved in the post-1980 epidemic, and why the eventual discovery and existence of subgroups within 1M, centered in the Congo region, argues for a long duration of this group’s dispersion within humans in the Congo region starting well before the 1950s.
5The current accepted account for the emergence of 1M’s subgroups is that 1M spread among humans primarily in cities in the Congo throughout the early 20th Century, with most of the resulting genetic diversity resulting from neutral drift rather than natural selection. Holmes mentions “a series of local founder effects (Rambaut et at. 2001). Specifically, viral lineages were by chance exported to other localities from a source population in the Congo region of Africa,” which was the cause of my initially published, incorrect remarks on the geographic footprint of early spread of the virus. 6
In misunderstanding the accepted epidemiology of pre-1980s 1M, I was allowing a mental model of geographically remote lineages to short-circuit a more important intuition regarding the lack of recombination in 1M’s subgroups.7
What is important about the development of the several 1M subgroups that were discovered after 1980, is that they diverged under conditions where the virus was rare. Although early circulation and therefore mutation was limited to cities in the Congo region, the prevalence of the disease remained low — still only measuring at .25% in 1970 in a sampling of mothers-to-be in Kinshasa (up to 3% in 1980).8 This kept the earliest offshoots of the virus from either competing with each other for and within new hosts, nor of recombining. This is the relevance of the lack of promiscuity in the 1M subgroups before the “pandemic” era.
What matters, in terms of cultivating a great depth of evolutionary divergence almost without recombinations — a family of high “purity” subgroups — is not so much for the virus to enjoy geographic separation but for it to successfully replicate in populations which, however geographically defined, have a very low prevalence of infected individuals that are interacting sexually or via shared needles or transfusion.
This ensures that any given strain — eventually, subgroup — remains unlikely to co-infect anyone at all times, for however many decades this condition of low prevalence is maintained.
(This section, itself, now has its own “put differently” formulation, which may help make clear the argument being offered. This appears in the correction notice issued for this post.)
The pre-existing depth of 1M measured against the post-1980 era
An important feature briefly mentioned in the last post, worth repeating, is that as the clock has progressed in the modern, post-recognition HIV epidemic this diversity of clades has not expanded in proportion to the expansion in human infections. By definition, subgroups outside of B, predominantly found in the Congo region, are not a product of the global proliferation of B in the 1980s, which itself did not produce these new subtypes.9 They are sisters, not daughters, of B. This is a way of answering the question, “well, so what about the subgroups?” The so what is that they reveal the pre-existing depth of genetic differences by allowing comparison to the shallow scale of evolution which has occurred afterward.10
Thus the picture as of 2000, when comparing global subtype proportions to Africa’s, was primarily that the spread of the virus through gays and drug users outside of Africa had been a minor bonanza for subtype B (simply because it was removed from competition with other strains, leading to a so-called founder effect benefit), but this bonanza did not contribute to diversifying 1M — the diversity was in Africa, and came from outside of B and in the milieu of endemic Congo-region HIV-1Ms before detection to which B belongs.
Here I must add an edit, in that my argument above may be mis-expressed if the threshold for genetic differences defining subgroups has been relaxed over time.
A more distinctive way of putting the argument, perhaps, is simply that 1M spent “a lot” of time in humans in an era where, again, prevalence was so low (in its home base of the Congo region) as to make recombination between its eight of so subgroups relatively uncommon, and now has spent “a lot” of time in an era where prevalence in several communities (globally) is so high as to have brought on a great expansion and proliferation of recombinants. Thus, the “pure” pedigree of the pre-1980 genetic divergence of the 1M subgroups remains an indelible testament to the existence of a long era of spread in humans before the widespread outbreaks recognized as the “pandemic.”
Finally, again, regarding the question of “well who is to say that this genetic diversity in 1M is not the product of multiple transmissions from chimpanzees, via medical research or whatever — perhaps just one chimpanzee virus 1M progenitor leapt into Americans, but multiple ones into Africans,” the short answer is that there is too much difference between HIV-1Ms and anything in west-African-chimpanzees.11 Sorting the sequences of HIV and SIVs using computer-magic-programs finds that 1M bundles together as a distinct node, rather than its different branches falling into different spots of the closest relative SIV-CPZ. (By contrast, other groups of HIV, which are less interesting from the standpoint of the epidemic, do find themselves grouped in different parts of the SIV landscape, reflecting wholly separate primate-human crossovers.) From Holmes:12
A further point can be made regarding the fact that 1M is human-adapted in a way which other HIV families are not, and therefore it is implausible that the genetic diversity of 1M’s subgroups originated in chimpanzees. This argument was made in reply to a comment by “Sleazy E” and is reproduced in the footnotes.13
Retroviruses in perspective: HIV as an unremarkable discovery
A second point, not yet mentioned, is that lentiviruses are genetically relatable to genes which have incorporated into animal genomes — these same genomes, our genomes, are replete with endogenous retrovirus genes, such as that which codes for the protein for human placenta fusion, and within this landscape of endogenous genes there are in fact relatives of animal lentiviruses — HIV is merely one class of this group. As Holmes writes:14
Despite the surprise of its emergence, as well as its devastating effect on human populations, HIV is not unique. […]
The wide diversity of mammals that carry these viruses, as well as the existence of endogenous copies without infectious relatives, not only suggests that they are an ancient viral family, but that there has been a regular birth and death of viral lineages.
In other words the “surprise” of HIV in the late 20th Century truly distorted perception of how “novel” it likely was. It, like other viruses which could only be isolated once antibiotics had facilitated the technology of cell culturing, was merely a newly noticed entity, already rich with genetic diversity indicating an endemic presence in humans of some length of time.
Again, this is to delineate that so far as the question of HIV being introduced to humans from mid- or late-20th Century medical experiments, including Koprowski’s wide-flung polio vaccine trials or weird chimpanzee-human cocirculation treatments for Hepatic comas, it isn’t supported by the genetic record as observed after the 1980s, and stretches too far to explain the mundane (i.e., why we would have and discover a human retrovirus that is of such an ubiquitous and ancient genetic platform that its relatives are even incorporated into our own genomes).

Of course, the reader is free to disagree with me, to consider my use of work by present-day zoonati members Worobey, Holmes, and Rambaut to be disqualifying due to the “stain” of their influence; this is fine.
Either way, this particular newly-noticed entity, misunderstood to be a foreign intruder (or not, as the reader might have it), was also in this case a newly manifested entity — rather than accounting for a previously well-recognized disease, another common cold or cough or pneumonia, the bizarre and apocalyptic disease known as AIDS “became.”
A host in change
Yet the way in which the disease “became” suggested from the outset that novelty of human social and chemical behaviors, not novelty of the virus itself, is the most obvious cause of the “becoming.” AIDS affected particular people doing particular things, and the virus to a certain extent obscured the importance of this observation by carrying with it conceptual baggage that time has rendered obsolete.
How can a newly-observed viral disease be caused by an old virus? — If this was not obvious in 1980, it is more-so now, in light of expanded knowledge. Many viruses infect immune cells, becoming dormant within their genes for life, without causing immunodeficiency of the type seen with AIDS — therefore we should not assume that HIV in of itself must cause AIDS, rather that perhaps it simply can, under specific conditions.
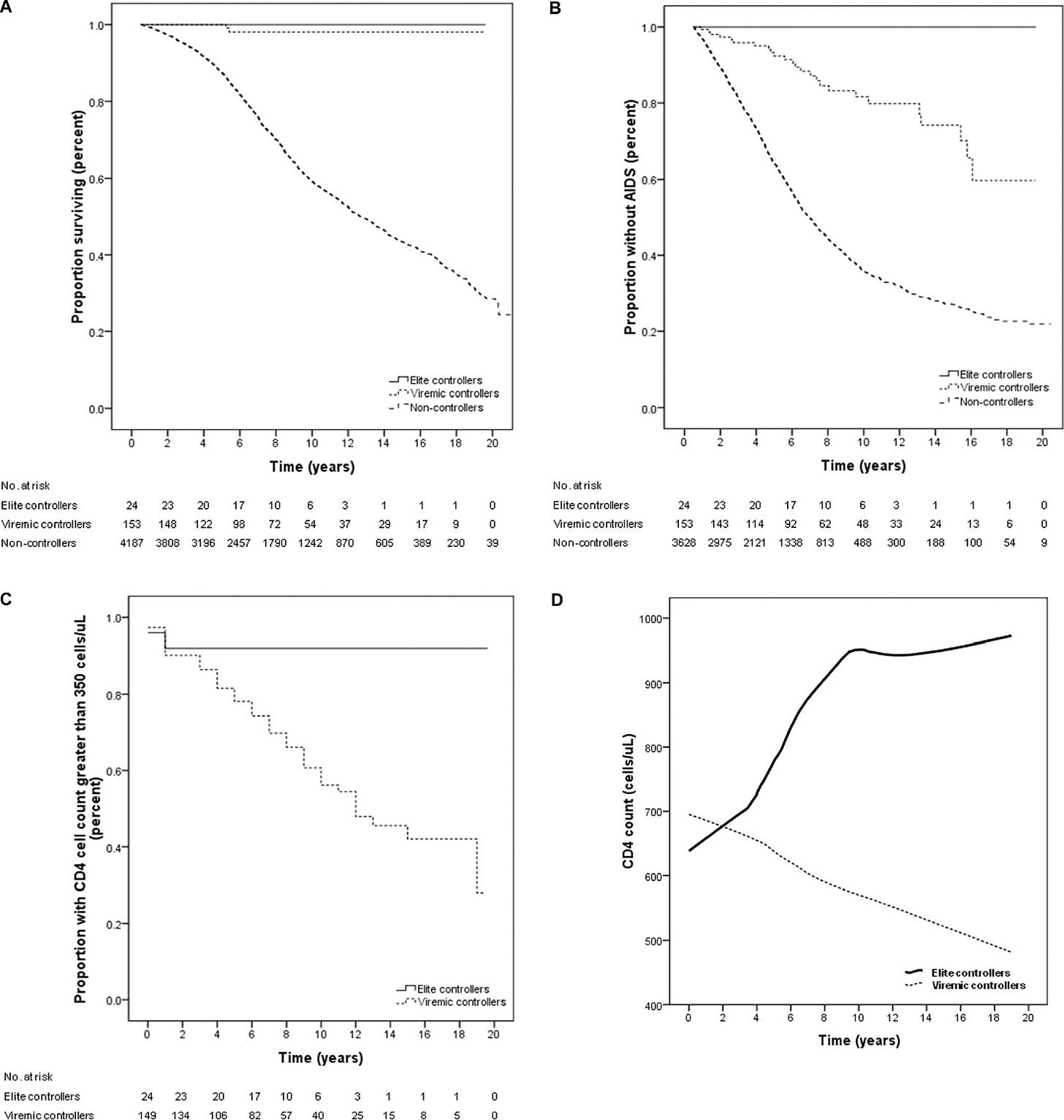
And this would seem to be consistent with varying outcomes of infection (even within gays alone), with some individuals being known as “long-term non-progressors,” (also “viremic controllers”) or “elite-controllers” (2.04 and .55%, respectively, of known HIV-1-infected military personnel observed for several years15). This phenomenon is observed without any obvious relation to genetic factors such as MHC phenotypes,16 but predominantly proceeds from early suppression of viremia, and is followed by low rates of AIDS and death.17 If the HIV “fire” is put out quickly, then it does not burn down the house.
Therefore the conditions of initial infection — the difference between a functional mucosal barrier and entry into the bloodstream — could be obviously of great importance. A counter-argument to this high-flying notion of a “safe” HIV infection is that most literature on HIV in Africa finds no great abundance of “controllers” among infected study subjects, and high rates of death.
The Hilleman vaccine as part of the change
At all events, we can put the theory regarding the First Hilleman Hepatitis B Vaccine into the category of potential answers immediately. Maybe the answer is because gay sex incurs trauma to the mucosa as well as immune insult from other STDs, and this gives the virus an advantage in early replication — so that infections result in a higher fraction of proviral T Cells. Maybe the answer is because needle-injection of the virus likewise circumvents mucosal immunity, and thus likewise results in higher initial replication. And maybe the answer is that Merck’s Hepatitis B vaccine injected people with HIV-infected plasma that wasn’t deactivated.
In other words the first thing to note about the FHHBV theory is that the cause of AIDS’s “becoming” is already over-determined. If HIV-1M was already endemic in Africa to any extent (which, it was), then the advent of intravenous drug use and un-repressed multi-partner gay sex networks outside of Africa are totally sufficient to explain why a normally unnoticeable virus suddenly proliferated in these communities and produced much more pathological infections.
So, did the Merck FHHBV vaccine amplify initial spread — both among American gays where the virus was previously unknown, and in Africa where the virus had simply been previously unnoticed?
The coincidence of the first vaccine based on the blood of (Hepatitis-B) infected individuals, and the explosion of HIV, seems meaningful — but again, this “coincidence” also applies to the timing of the vaccine and of these novel cultural behaviors. In fact the whole marketing rationale for the vaccine was that gays and drug users kept spreading Hepatitis B to each other — a problem that did not exist before the 70s.
Therefore the question has no prima facie obvious answer — the FHHBV isn’t “necessary” to have caused the outbreak of AIDS, at least among gays and drug users — and as such may simply be treated as a question of the related evidence.
If you derived value from this post, please drop a few coins in your fact-barista’s tip jar.
As well as a number of sub-groups comparable to known SIV viruses and to other endemic human viruses. The genetic diversity of HIV-1M is probably greater than that of influenza A subtypes in humans, though I haven’t done the math. Influenza A genes are either subject to strong purifying selection (stasis) or to frequent evolutionary sweeps, which culls genetic diversity* — but it is still useful as a touchstone for considering the depth of genetic differences in HIV-1M. It also seems to be greater or comparable to that for specific coronavirus types; however, these are more poorly sequenced than HIV or flu.
*Webster, RG. Bean, WJ. Gorman, OT. Chambers, TM. Kawaoka, Y. (1992.) “Evolution and Ecology of Influenza A Viruses.” Microbiol Rev. 1992 Mar;56(1):152-79.
Edward C. Holmes. The Evolution and Emergence of RNA Viruses (Oxford Series in Ecology and Evolution) (7.2.2). Kindle Edition.
Pan troglodytes troglodytes. All references to the same in the initial versions of these posts incorrectly used “western” instead of “central” to describe the relevant chimpanzee, due to my transposing of the Anglosphere convention of describing this part of Africa as “western.”
(Holmes), 7.2.4
The initial version of this post described early HIV as having spread throughout Africa, beyond just the Congo region, as noted in the correction. This gaff resulted from failure to encounter any references to archival investigations which found no evidence of the virus in a large number of samples throughout the rest of Africa before the 1980s, an omission which makes it hard not to mis-infer any summary description of the virus’s early dissemination. Also, I have something of a selective dyslexia that applies specifically to geography, frequently remembering the wrong placement of things as described, due I believe to a preference for wholistic thinking (see again the confusion with the designation of chimpanzees).
(Holmes), 7.2.3
This lack of recombination was at first a bit of an assumption by early researchers, as well as on my part while writing this post, as it is how the 1M subgroups are widely treated in the literature. It is however a perilous assumption, as phylogenetic trees are often constructed using incomplete stretches of the genome, which can preserve asexual lines of “pure” inheritance despite recombination somewhere outside of the same stretches. I have now found a study which checks the assumption by subjecting the subgroups to modern methods to look for recombination. One early recombination was reclassified as a pure subgroup, and one subgroup as a recombination (CRF), none of the other canonical subgroups were found to be CRFs.
Abecasis, AB. et al. (2007.) “Recombination Confounds the Early Evolutionary History of Human Immunodeficiency Virus Type 1: Subtype G Is a Circulating Recombinant Form.” J Virol. 2007 Aug;81(16):8543-51.
When using each subtype as query sequence against the remaining subtypes in a Simplot/bootscan/SlidingBayes analysis, we found clear indications for recombination only in subtype G
This figure is from Jaques Pépin’s extremely helpful text —
Pépin, Jacques. The Origins of AIDS (p. ). Cambridge University Press. Kindle Edition.
Which is responsible for identifying the mistakes in the original versions of these posts.
The specific citation is to
Desmyter J et al. Anti LAV-HTLV-III in Kinshasa mothers in 1970 and 1980. Presented at the International Conference on AIDS, Paris, 1985.
What has expanded, instead, are recombinations. The latter is a product of the global admixture of west African HIV viruses in co-infections that did not occur when the virus did not exist in high concentrations in any communities — it is a result of modern HIV-positive individuals being more likely to meet each other rather than simply pass the virus on to a previously negative individual.
Again, see previous footnote.
What such a situation would look like is west-African-chimpanzees literally being abundant today with human HIV (or some elusive “virus X” progenitor, but this isn’t really a meaningful distinction), rather than in a different but “closely”-related virus.
Thus, molecular estimates based on these larger data sets, carried out in the 2000s, put the divergence between “SIV-CPZ” and the ancestor of HIV-1M somewhere in the early 20th or mid-19th centuries, with SIV-CPZ having itself drifted from the cross-over virus in the intervening time (in other words, modern western-Chimp-SIV would first have needed to travel back in time to have originated HIV in the mid- or late-20th Century, based on changes in SIV-CPZ from related viruses, at least if the magic-computer-programs work the way I understand them to).
Of course, I have already mentioned that I consider these estimates to be “floors,” not ceilings, though I do not insist on the point.
As a sub-footnote, in fact they are treated as ceilings, but this ignores that if HIV is endemic in humans it will be subject, already and forever, to a bias toward reversions of deleterious mutations during transmission (as it is already understood to be, in present mutational study, see (Holmes), 7.2.5 & 7.2.7 — “Such reversions [of individually helpful intra-host mutations that aren’t beneficial for transmission] may also reduce the rate of nucleotide substitution at the inter- compared to the intra-host level (Maljkovic Berry et at. 2007)”) which leads to underestimation of mutational distance (because of failure to count unknown reverted mutations).
(Holmes), 7.2.2
https://unglossed.substack.com/p/the-hep-b-vax-hiv-origin-theory-pt-f53/comment/41225656
1, A Camp Lindi shenanigans theory offers an answer for the distance on the 1M tree but fails to satisfactorily explain the subgroups. Quite the opposite, you want geographic isolation [rather, you want prolonged low prevalence within an interacting group of hosts, as clarified in the edits to this post; the rest of the comment will contain some inappropriate statements based on the earlier idea, in strikethrough]
Then you have the tropism problem. HIV 1M has a very important adaptation to restore inhibition of human tetherin (the other HIV families don't), so it's human-adapted (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2779047/). If the subgroups of 1M developed in chimps then you wouldn't expect preservation of this adaptation, and certainly wouldn't be able to explain widespread adaptation across these diverse subgroups (in chimps or in human crossover infections) as opposed to just one that crosses over and dominates the globe (as the 1M group has done), given that HIV-1N/O and HIV-2 have failed to achieve the same adaptation to human tetherin despite being in actual humans.
So the 1M subgroups probably were grown in humans after 1Mancenstral gained this adaptation, it's once again not plausible that chimps nursed their genetic diversity.
Then you have the question of whether the answer RE distance is actually very good. Would there be effective "candle carrying" in such a situation or would you have, again a lot of loser infections which don't propagate their mutations, strains thus dying out, and new infections are for whatever reason from a minority of chimps that stay in the camp longer. Similar to how swine flu is antigenically very stable due to the annual slaughter of pigs, you can retard viral evolution when you move animals around unnaturally, rather than advancing it.
Then you have the question of why camp chimp strains of HIV fail to reeneter the wild chimp population. If they still have chimp tropism (despite also developing and keeping human tropism implausibly early and long in the case of 1M), then once humans start to spread them as well, they should go back into chimps, and then when we look at viruses in chimps we find these proto-1Ms in many of the different subgroups, because after all the claim is that 1M was grown in chimps, it would spread in chimps. As said in this post footnote 7, we would essentially just see HIV in chimps, and 1M would fall into different parts of the chimpanzee HIV tree. But we haven't found that.
(Holmes.), 7.2.1
Okulicz, J. F. et al. “Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV Natural History Study.” J Infect Dis. 200, 1714–1723 (2009).
Díes-Fuertes, F. et al. “Transcriptome Sequencing of Peripheral Blood Mononuclear Cells from Elite Controller-Long Term Non Progressors.” Sci Rep. 2019 Oct 3;9(1):14265.
(Okulicz, et al.)






HIV was present in the Congo in 1959
https://pubmed.ncbi.nlm.nih.gov/9468138/
HIV infection in a 16 yo in St Louis in 1968
https://jamanetwork.com/journals/jama/article-abstract/374422
HIV was present before the homosexual revolution, gay bars and bath houses. It was not widely spread until the change in social norms.
Greetings Brian - long time no see! Why? Because my email started sending your posts to spam beginning July 27. All this time I thought you were taking a break! 😠 Did ya miss me? 😄 Oh well, plenty of good stuff to read while doing the laundry.