The author is recovering from apparent mild food poisoning today. It’s nothing horrible; and I was able to whip up a demonstration I have meant to make for a while. In which, the “gist” of how sequence-based phylogenetics works is demonstrated in a bare-bones cartoonified fashion.
This post regrettably still comes attached with an Out of Office notice — I will be away from my workstation until Tuesday (food poisoning permitting).
Context: Launching a case study series on transmission dynamics
This guide seeks to ground readers on what we can infer from collections of sequences. Do they support the case that SARS-CoV-2 is transmission-competent, self-sustaining virus (meaning that any claims otherwise must allege widespread sequence falsification) — or not?
If you are wondering how, in the chaos of millions of uploaded genomes over the last 3 years, there could be any chance of making sense of anything — well, that is exactly what this post hopes to illustrate.
To be clear, I am open to the claim that SARS-CoV-2 cannot sustain itself in human-to-human transmission; that only “vector-delivered” versions of the virus can generate severe symptoms, and that the virus must constantly be re-injected into human transmission by some means.
The only difference between my position and that proposed by JJ Couey in this respect is that I do not believe this is necessarily true, i.e. due to some hidden intrinsic property of all RNA viruses. In other words, I think this might be a feature of this virus, despite not being a feature of long-adapted, co-evolved human viruses.
Why human RNA viruses are not all “traveling salesmen” from a zoonotic reservoir, doomed to fade out after a few infections
If RNA viruses all self-limited, why would we bother making lifelong immunity to any of them (antibodies, including life-long plasma cells (LLPC1); B and T Cell tissue deployment; innate antiviral reprogramming; etc.)?
Wouldn’t that be futile if the immune system didn’t expect RNA viruses as a default to (from the cellular perspective) “come back later”?
In the wake of my extravagantly failed attempt to convict the 2020 VOCs of the crime of being lab-made mutants, my version of this project involves a long, slow odyssey through early and late dynamics of sequences. I expect, based on previous poking-around, to notice three “phases”:
But if I find evidence of artificial transmission and mutation in all three (or none), so be it.
This approach will be unusual in employing no large scale phylogenetic computer modeling. My only comment is that such algorithms are not designed to detect artificial release; they must default to finding either nothing or to showing patterns (i.e. natural transmission) that aren’t really there; and no one has any way to audit the results. Therefore, I will focus on a “spot checking” approach to see what inferences really hold up to the evidence on the small scale.
With that context out of the way, here is a “splainer” for what sequences can and can’t tell us about transmission dynamics.
Cartoonified Phylogenetics 101
i. Sorting save files, the easy way
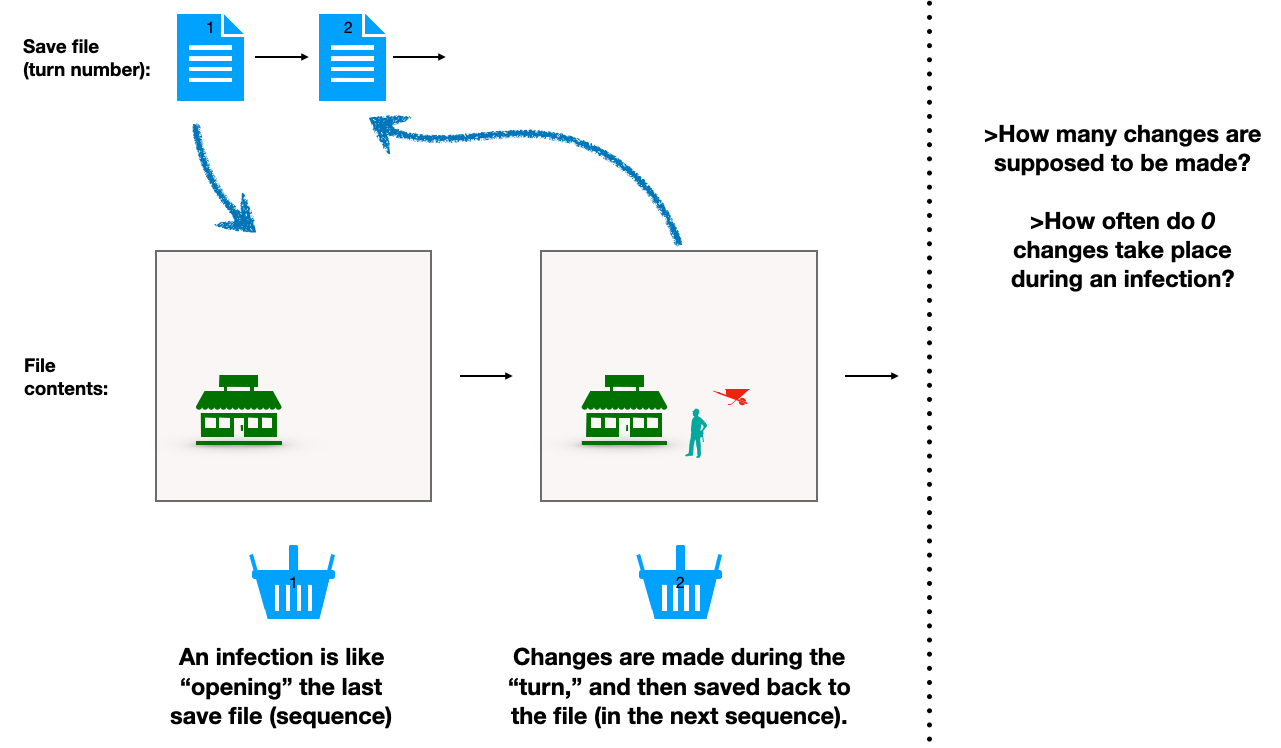
First, imagine that you have bought a turn-based strategy game.
In each turn, you are allowed a set number of actions. At the end, you hit “next turn,” at which point an auto-save file of your current map is saved, with said map reflecting the changes made in the just-completed turn as well as whatever was already lying around from prior turns.
The result is a sequence of files that you can re-open later, to see the snapshot of your map at the end of each turn.
You know that the file labeled for turn 3 is going to show the changes made on turn 3; in addition to however the map already looked on turn 2.
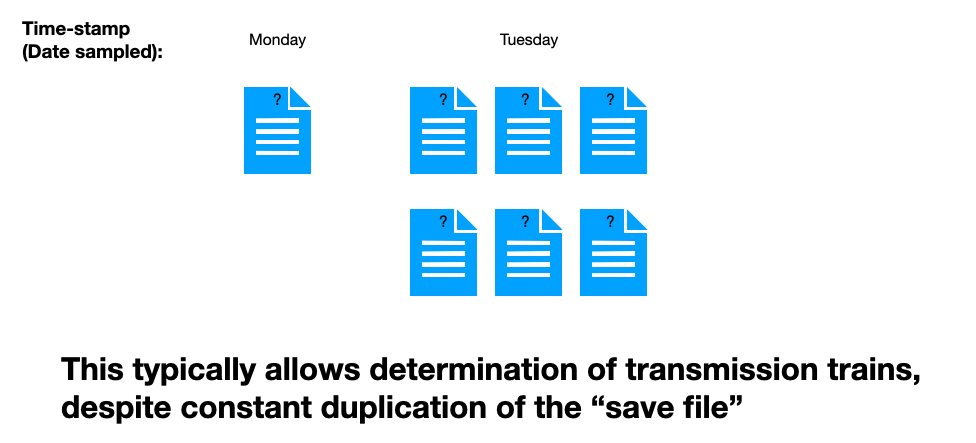
But what if your save files are not labeled by turn number? What if you only know “other stuff”?
Which “other stuff” in this slide will tell you the turn order of the files? It isn’t the time-stamp, obviously. But you still have every chance of figuring out the correct order. The “information” that provides that missing order are the maps described by the save files themselves.
You know (given these rigid assumptions, and perhaps a requirement that all changes besides moves are permanent) that the middle file came after the leftmost file, which itself came after the rightmost file — changes must grow in time. (If you have played these types of games, of course, you can actually prove this in practice with blinded turn number loads.)
ii. Extending to a virus
This is the same process that allows figuring out the order of mutations that lead SARS-CoV-2 to “grow changes” from its established baseline sequence.
We’ll skip over how the “oldest” or baseline sequence comes to be known in most cases. With SARS-CoV-2 it was suspiciously determined absolutely by sequencing a single patient (“Wuhan-Hu-1”). This turned out not to be the true baseline; but that isn’t important either. Wuhan-Hu-1 has served well in practice (suspiciously so).
When comparing two sequences, a progression can be inferred by shared and unique mutations. They might have no apparent relation relative to the baseline (i.e., they are distinct strains) or they might clearly stem from a single lineage.
This should be how transmission is modeled in discrete outbreaks. We ought to be able to look at the basket of captured, higher-quality sequences collected during an outbreak and sort the whole story out. Again the first step is to take our basket of sequences —
— and then open them. i.e., use the “information” stored in the genes themselves (growing mutations over time) to figure out the story:
Here, the index case is presumed to have infected three groups of people. But is this actually a mandatory conclusion? Not at all. Because in this case, there are numerous sequences without any apparent mutations. No mutations, means no information.
iii. The mystery of consistency in SARS-CoV-2
But this shouldn’t be a big problem, because “everyone knows viruses mutate all the time,” right?
But with SARS-CoV-2, especially in the earliest days, it proves to be a tremendous problem. In a preview of the first case series we will tackle, here is an early report from a series of “outbreaks” in Austria:

Nearly half of these quality-benchmark-meeting sequences are “clones” in the literal sense — they have identical genomes to other sequences in the same cluster. These same “clones” further bear almost no sign of “traveling to Austria” in the first place. B.1 and B.1.1 together comprise the “D614G” siblings that give rise to all future strains; they seem to arise in Italy (B.1) and the Middle East (B.1.1) in mid-winter, before the virus is even announced2 — just how is it that they have come all the way from their original emergence to Austria, months later, with only 1 or 2 extra mutations?
These genetic curiosities are the basis for the phylogentic inference that the virus tends to infect most people via “super-spreaders.” It goes without saying that this conclusion was reached without considering other means of “spreading” nearly-pure versions of B.1 to a large group of people all at once.
Moreover, the conclusion that super-spreading explains these patterns begs the question; because without (growing) mutations, we do not have information. To repeat the previous slide:
We can assume, or train computers to declare, that all those identical sequences are the result of a “super-spreader.” But because those sequences are identical, by definition, our assumption is based on nothing.
The key question to help resolve this problem is, how many changes (mutations) is SARS-CoV-2 supposed to make, per infection / transmission? Well, we don’t really know.
Thus, when it comes to the belief that super-spreading is a property of the virus, that depends in large part on an assumption that infections should add changes (mutations) almost always. But it’s just that, an assumption.
The lack of information in identical (i.e. clonal) sequences is thus itself a form of information. We simply have no reference to discern just what that information is.
Does it mean that single individuals really do typically infect dozens, if not hundreds, of others? Or does it mean that “pure” genomes are being distributed by other means? Or does it simply mean that SARS-CoV-2 (at least at first) bore an intrinsically ultra-stable genome that hides half of the actual transmission chains in early outbreaks?
This is what this case study series will explore.
Related:
If you derived value from this post, please drop a few coins in your fact-barista’s tip jar.
The LL is for “long-lived,” but I am specifying how long; it happens to make the same abbreviation.
Amendola, A. et al. “Molecular evidence for SARS-CoV-2 in samples collected from patients with morbilliform eruptions since late 2019 in Lombardy, northern Italy.” Environ Res. 2022 Aug 25;215(Pt 1):113979.
Further discussed in “The Quiet Corona-Can.”



















That's a great post and I am struggling with the same questions. How many of the "variants" were lab designed vs naturally evolved. The initial Omicrons were possibly designed around Jul-Aug of 2021.
All right, I'm really glad you're writing this series. I try not to miss a live stream with JJ Couey.
I hope you feel fully healthy again in no time. ❤️ Food poisoning can be awful!