Avian flu outbreaks, Genetic code map (some media uploads)
A flu-timeline-work-in-progress post.
The sole purpose of this post is to offload two media resources that might or might not be informative or curiosity-striking to the reader. These may be republished in Part 2 of my “OAS lit. review,” or perhaps a stand-alone treatise/“explainer” on the genetics of flu.
In the meanwhile, tomorrow begins a final 5-day offline hiatus, after which this journal’s rate of posting will hopefully return to pre-summer levels.
Although both items are presented here with only the barest of user guidance, highly curious readers may find the material contained within either obvious or intriguing, depending on level of familiarity with flu and genetics. Whereas readers currently in “have a life” or “summer-chill” mode should probably stop scrolling here.
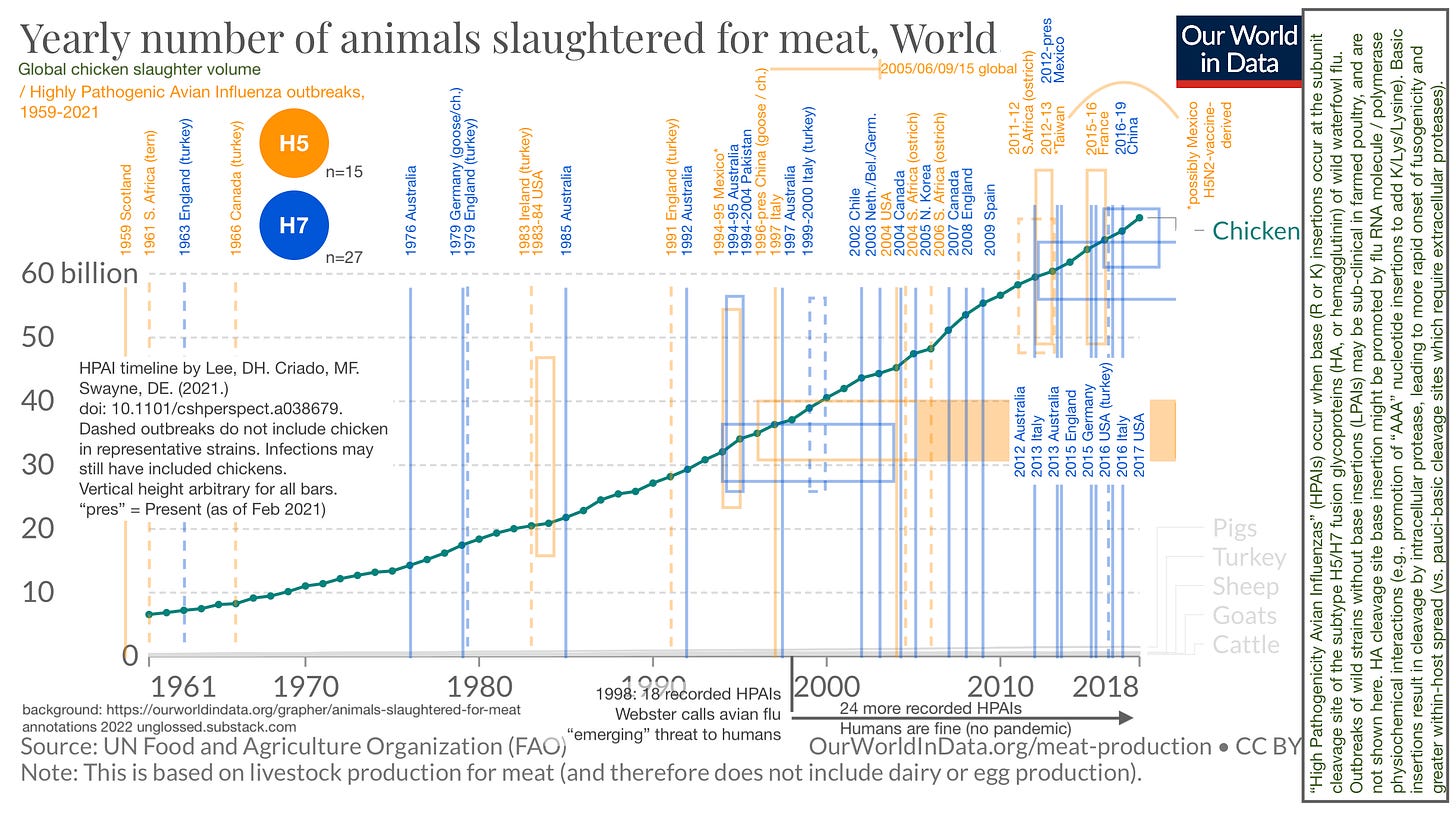
Resource 1: Highly Pathogenic Avian Influenza outbreaks, 1959-2021
It turns out, human flus derive from avian flus. This won’t be news to anyone who has been obsessing over or even paying slight attention to (theoretical future) flu pandemics since 1998; but for me, having always been skeptical of every aspect of flu research, it required reviewing the history of the field from the very beginning to grant even the first ounce of credence to any of the modern work. And, wow: Avian flu is really where it’s at.
Here, I have done my alleged best to visualize the increase in recorded HPAI outbreaks in relation to the linear increase in chicken consumption (which is the almost exclusive driver of gains in overall meat consumption since the mid-20th Century).1 This graphic also serves to demonstrate that the “match” of H5/H7-based HPAI outbreaks has been struck so many times, without consequence, that spillover into a bona fide human influenza “pandemic” should probably be considered unlikely at this point.
Resource 2: Brian’s Crazy 2/1/3-Codon Genetic Code Map
Formerly designated “CUGA,” when first published here in February.2 This map positions the residues (and M/start and “nonsense”/stop commands) of the genetic code used by all life-forms by the second codon-letter, on the (totally speculative) premise that this letter determines ribosome/tRNA synthesis rate and therefor serves as biology’s preferred method of “grouping” non-radical residue changes (in other words, since changes to the 2nd nucleotide/letter are likely to be radical in residue type, they are probably radical in RNA read-dynamics as well).
The version of this map published in February positioned G and A at diagonals from their pyrimidine mates. This version corrects that.
Again, this post is not intended to provide a full user guide to the media in question. But the “elevator pitch” is that this “map” displays how changes to the second letter result in radical alterations of residue properties; changes to first letter result in “compatible” - or radical, but potentially contextually highly likely to be beneficial alterations; and changes to third letter are the least significant (often being synonymous), perhaps due to lower read accuracy as is commonly speculated. It is lastly suggested that G and A (purines) in the 2nd codon-letter decrease read speed and increase read-accuracy vs. U and C (as inferred by co-localization of the stop codons in the G and A quadrants).
Regarding “radical, but potentially highly likely to be beneficial” for 1st-letter-changes, an example in the context of influenza is the “PB2” gene/protein in the polymerase triplet. A mutation from E to K involves switching the first letter of the codon for E from A to G and toggles tropism of the virus as a whole from birds to mammals:3
A single amino acid change in PB2, E627K, was shown (1) to be important for mammalian adaptation2,3, (2) to distinguish highly pathogenic avian influenza (HPAI) H5N1 viruses in mice19, and (3) to be present in the single fatal human infection during the HPAI H7N7 outbreak in the Netherlands in 2003 (ref. 20), and in some recent H5N1 isolates from humans in Vietnam and Thailand and wild birds in China21,22,23.
Accordingly, the 2/1/3 “map” demonstrates that an E to K change both reverses charge, maintains size, and only requires a nucleotide swap in the 1st letter.
Accident, or design?
Suggestions for a new name for my map are welcome.
If this extremely user-unfriendly post has left you consumed with the rage of four thousand exploding suns, please drop a few coins in your fact-barista’s tip jar.
Sources:
https://ourworldindata.org/grapher/animals-slaughtered-for-meat
Lee, DH. Criado, MF. Swayne, DE. “Pathobiological Origins and Evolutionary History of Highly Pathogenic Avian Influenza Viruses.” Cold Spring Harb Perspect Med. 2021 Feb 1;11(2):a038679.
(And a link about cleavage site nucleotide insertion-promotion that I need to dig back up.)
See “Upside Down Fractal CUGA,” which attempts to provide a walkthrough of how to visualize mutations, although most images still use the switched A/G positioning.
Taubenberger, JK. et al. (2015.) “Characterization of the 1918 influenza virus polymerase genes.” Nature. 2005 Oct 6;437(7060):889-93.