Shaky DNA integration study
Merely designing a study to answer this question is a demanding task - here, it is not quite met.
Igor Chudov continues to produce excellent work and maintain his position on the leading edge of interesting developments this month, and so once again I am following in his footsteps - though as usual, with that ambivalent, Unglossed-style reading of the evidence!
Today, he highlights a paper purporting to demonstrate in vitro DNA integration after mRNA transfection with the Pfizer/BioNTech Covid vaccines.1 Obviously, such a paper would be answering a question that is on the minds of many.2
However, I think the study does not offer a very satisfactory answer to that question. Even if DNA integration is occurring in real-life injection, this paper doesn’t add evidence that improves upon the theoretical argument that is already available. It wasn’t designed in a way that would robustly demonstrate that effect, and there are quality signals in the results. I will be brief and non-comprehensive, since the goal here is to demonstrate a negative. Readers who want a break-down of the study design and purported findings first, should head to Chudov’s post and then come back.
The problems:
Not set up to demonstrate the effect.
Total flunking on the LINE-1 expression mechanism.
More weird results with the controls during chaperone protein localization.
Provisionally rescinded: Red flag for false positives on the headline results.
In summary, studying DNA integration requires very precise and robust study designs. This paper set out to accomplish something ambitious without an ambitious approach.
Not set up to demonstrate the effect.
Beyond being in vitro, so that any findings would be only be a precursor to further, real-world testing, the study was based on Huh7 liver cells. Like many other cell lines used in virus research, these are immortal cancer cells, which carry around ~60 chromosomes full of erroneous insertions and deletions to begin with:3

Is this really a good proxy for how non-diseased cells would respond to foreign mRNA, or the downstream metabolic disruption induced by producing an unknown amount of copies of a foreign protein for an unknown length of time? The authors should have verified their methods by demonstrating the performance of another cell line under similar conditions. Indeed, we will see some weird behavior in the untreated (not-mRNA-dosed) Huh7 controls below.
Again, there are plenty of theoretical arguments already available for how foreign mRNA designed to evade normal break-down could obviously carry an increased risk of insinuating into DNA - compared, for example, to mRNA that was designed to degrade quickly. We should obviously be concerned. But how does putting this mRNA into these poor little Akira liver cells that someone needs to just let die help confirm or falsify those concerns?
Further, the authors used un-dosed “controls.” Since they were trying to show a four-part mechanism - exposure to the treatment, effect on LINE-1 gene expression, accumulation of LINE-1 chaperone protein (the protein that takes LINE-1 mRNA, and sometimes foreign genes, back into the nucleus for eventual copy-pasting into new parts of the genome), and Pfizer spike DNA (implying integration of the mRNA), failure to control for the other ingredients of the product makes it harder to attribute vs.-control differences in the other mechanisms to the mRNA rather than to the other stuff. So, it would probably be a good idea to have included “blank” (as in, coding for another protein) mRNA- and LNP-dosed “controls.”
(Edit, March 5:4 Thanks to TJ Lees for pointing out, in the comments, that since the Pfizer formulation is proprietary, the authors didn’t have any way to meet the bare minimum treatment-control proposals suggested here. So, in a comparable study design without a proprietary product, a reasonable design would be, “Plasmid for the spike,” “Plasmid for a Influenza HA protein,” for example; but in this case “Pfizer / BioNTech LNP for HA” is not feasible. However, they still should have used multiple cell lines.)
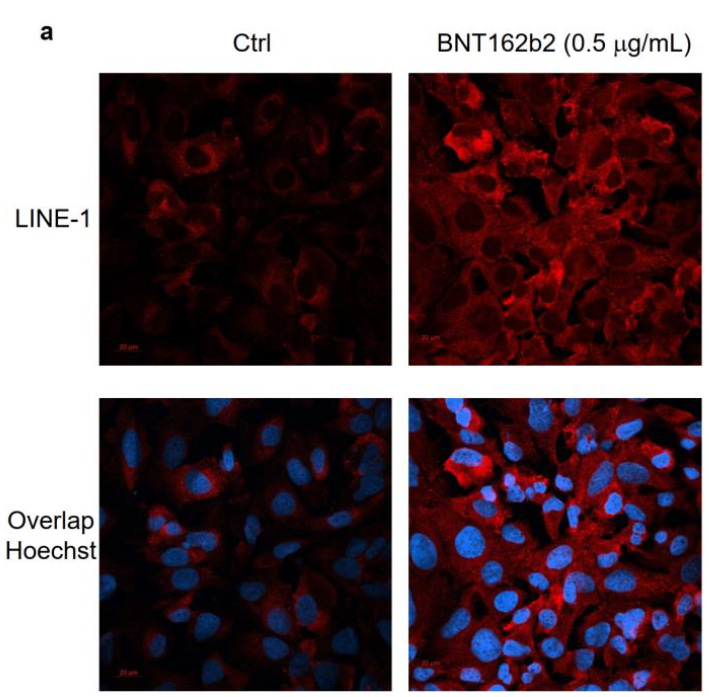
Total flunking on the LINE-1 expression mechanism
Again, this was bound to be a weak finding anyway, with such a skeletal study design. But note how the control LINE-1 expression apparently goes haywire as time goes on. In fact, it seems like the treatment is suppressing the “normal” (as in, abnormal due to cancer) LINE-1 expression of the Huh7 cells, but that high dosage elicits a confounding, smaller, pro-LINE-1 effect:
They really probably shouldn’t have used cancer cells. Especially not the “highly heterogeneous” kind.
What the authors are attempting to show here is that the treatment is stimulating transfected cells to shoot out more of the LINE-1 mRNA that leads down the path toward DNA integration. By comparing vs. housekeeping gene mRNA, they may be accidentally measuring a difference in expression in those genes.
Again, it’s not clear how a strong signal here would demonstrate mechanism without more cross-controls anyway.
More weird results with the controls during chaperone protein localization.
For some reason, the untreated control nucleuses don’t stain as brightly in Fig 4. Did the authors simply mess something up with their controls, or the exposure of the stain? It’s impossible to tell. In Fig 4a, the control blue areas should be just as bright as the treatment blues, in order to compare the red areas (which correspond to Orf1p, the protein coded by LINE-1 that binds to mRNA and guides it back into the nucleus).
Fig 4b seems to compensate for this - but we can’t know how much chaperone protein signal was dropped due to the apparent low exposure.
Provisionally rescinded: Red flag for false positives in the headline results.
Here, the authors couldn't make “controls” 5 or 6 show primer-targeted Pfizer gene sequences:
My original comments on this have been archived to the footnotes, thanks to the following feedback:5
Edit, February 26: Modern Discontent, who has done a sequence or two, offers feedback in the comments that the language surrounding control 5 and 6 lacks clarity but doesn’t seem to suggest control 6 should be a positive.
I think the analysis of the gel may be incorrect. It appears that Ctrl 5 and 6 were done to show that their amplicon had to have been arrived from DNA, such that Ctrl 5 and 6 essentially did an RNA extraction. Because both did not have any RNA, no amplicon was found. I think their wording made it confusing. They should not have said they performed PCR on RNA purified from the cells but that they did an RNA extraction to see if they could find any RNA to begin with.
Edit, February 28: TJ Lees, who has performed the relevant steps in this study, provides more push-back in the comments, and offers an explanation for the authors’ RNAse yes/no control design that strikes me as plausible, though superfluous in some respects.6 Moreover, Lees corrects my implicit misunderstanding that PCR (not “RT-PCR”) would be likely to amplify RNA by default, without a deliberate additional reverse transcription (“RT-”) step.
Lees also objects to my gel comparison (hence the disclaimer immediately below), and the point may be fair. (Lastly, Lees offers a plausible non-LINE-1 mechanism for reverse transcription that may make the 6 hour results more reasonable: The cells may have been contaminated with a retrovirus to begin with.)
Disclaimer: Provisionally demoted to “working thoughts” status, reader to use own judgement:
There should also probably be more hits in the 48 hour group, if they are measuring integrated DNA and not RNA from the vaccine. LINE-1 protein-bound mRNAs perform integration during cellular division; often after a cellular division has already occurred and prompted the migration into the nucleus.
Instead, levels of tagged Pfizer genes drop between 24 and 48 hours - as if, what the authors are actually measuring as “integrated DNA,” is just mRNA from the treatment (though, again, the lighter “L” imprints in the 48 hour group imply a lower “exposure,” so the amount of Pfizer genes might actually be comparable to 24 hours):
Really, if the authors’ LINE-1-centric mechanism is in fact taking place, these blots should show almost nothing at 6 hours, then more at the other two. So, again, the results don't support the proposed mechanism very well.
To reprise the introduction, it wouldn’t surprise me if what this study alleges it is showing is happening in real life. But the design is a mess; it probably should have been rejected in peer review.7
We’re better off with theory, instinct, and common-sense caution, for now.
When I posted my comments on Chudov’s post, I received the following request from reader “Susanna”:
Could you comment on the theoretical comparison of vaccination versus natural infection potential mechanism of DNA integration?
Perhaps the same thought might occur to readers here (I should be so flattered). Here was my somewhat hedge-y reply:
I think there's no reason to rule out either without well set-up studies. The virus destroys the cells it gets into (there doesn't seem to be a lot of evidence that SC2 goes dormant like chicken pox or other viruses, nor reason to expect it would). The mRNA transfections ("vaccines") don't, so that's a huge theoretical skew in the risk for DNA integration and harms via metabolic disruption - both of which can get you to cancer.
In other words, the virus doesn't leave any "cell" to integrate DNA into. There might be some integration via marginal dormancy or intracellular-suppression, but maybe DNA error checking sorts that out. Maybe this is a common thing with all infections. We don't know the baseline here. Our genes change throughout life, except in the "snapshot" saved in our germ cells, but those changes can be more thought of as "yearbook signatures" rather than rewriting the text of the yearbook.
Though I am personally less worried about the “integration” question, since I take altering our gene expression to warrant concern over all same the ethical and medical hazards as altering our genes themselves.
Edit, February 26: The original version of this post was overly sensationalist with the Huh7 cells. I have modified the text here to be a bit more even-handed. Original text:
Beyond being in vitro, so that any findings would be only be a precursor to further, real-world testing, the study was based on Huh7 cells. These are immortal cancer liver cells, which carry around ~60 chromosomes full of erroneous insertions and deletions to begin with:
This is is not a good proxy for how non-diseased cells would respond to foreign mRNA, nor downstream metabolic disruption induced by producing an unknown amount of copies of a foreign protein for an unknown length of time.
I have left in the Akira reference, as it is merely descriptive of what cell that once belonged to a human and is made to divide over and over and undergo billions of genetic manipulations would call its “life experience.”
I should have added this edit in the earlier round, hence the late timestamp. In the preceding paragraph, the reference to “blank” mRNA was also clarified.
The following original text was removed:
It should have been, 6 shows amplicons, 5 doesn't, to confirm that their RNase treatment is protecting the DNA results from false positives. So the DNA results might be... false positives. As the authors put it:
To ensure that the DNA amplicons were derived from DNA but not BNT162b2 RNA, we also performed PCR on RNA purified from Huh7 cells treated with 0.5 µg/mL BNT162b2 for 6 h, with or without RNase treatment (Ctrl 5 and 6 in Figure 5), and no amplicon was detected in the RNA samples subjected to PCR.
If the authors weren't intending to get a positive on control 6, it's not clear how this was "ensuring" anything.
In either Modern Discontent or TJ Lees’ explanation for the double-control design, only one control actually ends up being necessary to flag the DNA amplification results via a positive; the other control may add information but is superfluous as it would not rescue the main results from a positive in the “necessary” control.
But the authors did not use just one control. They used two.
However, my interpretation was also based in part on an overly loose understanding of the term “PCR.” (“PCR” usually excludes “RT-” (reverse transcriptase) when used in a research context, whereas I imagined that the “RT-” step was functionally built in. Even in published research, “RT-” will sometimes be subtracted when describing amplification of mRNA. However, in this paper’s methods section, “RT-PCR” is explicitly referenced for the gene expression portion and only “PCR” for the amplification / gel portion.)
Interestingly, this study took place geographically- and publisher-adjacent to another study about which I have reservations due to its “over-ambitious” design, and over-reliance on staining. This is the infamous “spike goes to the nucleus and does stuff!” study. Here, again, the staining images didn’t really seem to support a strong conclusion. See Jiang, H. and Mei, Y. “SARS–CoV–2 Spike Impairs DNA Damage Repair and Inhibits V(D)J Recombination In Vitro.”
To me, it doesn’t look like the spike is in the nucleus. Maybe just “in front” of it a bit. Cameras are two-dimensional, after all. But, as the design wasn’t as over the top as this new paper, I did not think it merited a post.









It is most incredible to have disagreements on substack and your article is very interesting. Thanks for discussing it. I am also baffled by LINE-1 expression not matching what I would have expected.
As someone who has used all the methods reported in this paper in various contexts, here are my views:
1) Choice of cell line. Cell and molecular biology studies use immortalised cell lines, i.e. those derived from cancer, all the time mainly because they are easy to grow in the lab. A study with primary cells (directly derived from an animal) or an animal model would be much harder to do and would have taken a lot longer. The reason for choosing Huh7 over other cell lines was noted to because of their hepatic lineage, but also was likely because the researchers simply had easy access to those cells; the latter is not a great reason, but reflects the simple reality that most studies like this make such choices due to simple practicalities. Results from cell lines obviously are not necessarily indicative of what would happen in other cells, but are usually the first step in such a study. They should, however, have performed the experiments in multiple cell lines, to improve confidence in these results.
2) Despite the use of a poor model, it does still show that BNT162b2 RNA can be reverse transcribed in vitro in human cells. This is still an improvement over the "theoretical argument that is already available" because theoretical arguments are still theoretical, and experimental evidence is needed to prove this. Despite its flaws, the study provides the first evidence that reverse transcription of BNT162b2 RNA in human cells. This is presumably due to endogenous LINE-1 (as they didn't add any retrovirus), but the evidence that LINE-1 was responsible is circumstantial at best; the data showing induction of LINE-1 is also poor. In addition, the study doesn't itself rule out the presence of retrovirus infection in these cells, which is another potential flaw in the study.
3) There was no realistic way to obtain "blank" mRNA or LNP controls. Pfizer would not have provided these, and any substitutions for "similar" reagents would erode their usefulness as controls. In addition, such controls would be interesting, but not strictly necessary. Even if the effects are occurring due to "other ingredients of the product", the whole product is what is being injected, and the effects don't need to be due to the mRNA itself. (Obviously, the LNP plus mRNA combination is needed to achieve anything, as the mRNA wouldn't get into the cells without the LNP).
4) Figure 3 is a mess but if you look at the text, they don't actually refer to the cross time-point comparisons ("Significantly increased LINE-1 expression compared to control was observed at 6 h by 2.0 µg/mL BNT162b2, while lower BNT162b2 concentrations decreased LINE-1 expression at all time points (Figure 3)."). They should have removed the statistical comparisons from Figure 3. The criticism about the induction of LINE-1 in the Ctrl at 48 h is valid but is not too surprising - culturing cells easily changes the expression of various genes. Criticism that this could be due to normalisation by housekeepers is also valid, but this is inherit in the vast majority qPCR gene expression designs, which almost invariably use housekeeper normalisation.
5) The differences in fluorescence intensity in Figure 4 do indicate that any quantification of such images is a problem. I'm not too surprised by this - obtaining consistent labelling intensity across different samples using fluorescence microscopy is extremely difficult in practice. Therefore, trying to quantified fluorescence intensity from such images is always a somewhat dubious method. Doing this "properly" generally requires addition of some kind of labelling control on the slide to normalise to, but this is usually not possible for most targets/experimental designs. Nevertheless, this method is used a lot in such studies, despite known issues, so this is no worse there thousands of other cell biology papers doing so. In addition, the researchers appear to have done a flawed statistical analysis for Figure 5 - they have used n=15 cells from two images, and quantified the fluorescence from this n=30. This is an example of pseudo-replication. The cells are NOT proper replicates as treatments are being applied to the wells of cells, not individual cells, and thus the correct level of replication needs to be at the level of wells. Yet, it seems that there was only n=1 well for each condition (for which two images were taken), which is extremely poor. It should be noted, however, that although most cell biology researchers would see the flaw in doing only n=1 well/experiment, most still incorrectly do statistical analysis on the pseudo-replicates of cells rather than wells, reflecting a poor level of training and understanding in statistics in the field (proper analysis of such a design involves treating it as a nested model, which is beyond the capabilities of most cell biologists).
6) I can't see why Ctrl 5/6 are a "red flag". Ctrl 5 and 6 seem to be exactly as stated: RNA extractions with and without RNAse. Both a negative controls, and they lack a positive control in those images. An appropriate positive control would have been RNA/no RNAse with a reverse transcription step before running the PCR. Nevertheless, Ctrl 5 and 6 seem to serve their purpose and are meant to be interpreted together. Ctrl5 (RNA/no RNAse) would indicate whether the PCR amplification occurred from an RNA extract, i.e. the PCR doesn't amplify RNA. As expected, there is no band here, as PCR will not amplify RNA (prior reverse transcription is needed convert to RNA). If Ctrl 5 had been positive, however, it would not be clear that this is due to PCR amplification of RNA instead of DNA contamination of the RNA. This is what Ctrl 6 (RNA/RNAse) is for. If Ctrl 5 had been positive (it wasn't), a positive Ctrl 6 sample would have shown that the Ctrl 5 band was likely due to DNA contaminating the RNA extract (as any RNA would have been degraded by the RNAse). If Ctrl 6 had been negative, it would have shown that the Ctrl5/6 bands were somehow due to amplification of RNA (but they were negative). Ctrl6 also serves to control for the presence of contaminating DNA in the RNAse, which would have been another possible source of a band in the other lanes.
7) Gels of PCR bands are at best semi-quantitative due to a variety of issues (which I can expand on if anyone wants me to), and therefore, in my view, it doesn't make sense to infer much from the band intensities in Figure 5. In addition, the three different images suggest three different gels. Comparing band intensities across gels is a huge no-no. The normal way to interpret these gels is simply the presence or otherwise of bands, which is exactly what the authors did.
In conclusion, I'd say this isn't a great study, and has some clear flaws in it, but it still is the first experimental evidence that BNT162b2 RNA can be reverse transcribed in vitro in human cells. This evidence is limited in that it is only in one cell line, and evidence linking this to LINE-1 is circumstantial and weak. Clearly more studies need to be done on this but (1) I doubt anyone is in a hurry to fund or perform such studies, given the prevailing narrative surrounding these vaccines, which makes questioning them a "cancellable" offence even in the scientific community and (2) such things take time. The flaws in the present study likely reflect the simply practicalities of doing such research when no one wants it do be done.
Really, detailed studies checking for the possibility of endogenous reverse transcription of the BNT162b2 RNA in human cells, and the possibility of genomic integration, should have been demanded from Pfizer prior to approval, so this study also serves to highlight what might have been found if these had been done.