All SARS-CoV-2 clades fade out
A "SVAH expansion pack": Variants of concern are not intrinsically more fit, and both pre-VOCs and VOCs seem to lose growing-power over time.
A follow-up to the Should variants actually happen? series.
Brian makes the case for the “Variants of Concern” not being uniformly more fit (so, fitness does not result from nor explain their simultaneous, seismic bursts of “experimental” mutation); and makes a video of all the variants fading out over time (just skip the first part and scroll to the bottom!).
v. Addendum Myth: The “Variants of Concern” were more intrinsically fit
Markael Luterra challenged my focus on adaptive immune evasion in the comments of yesterday’s post, proposing, instead, that the variants might putatively be explained by innate immune evasion or gains in infectiousness; both of which I would characterize as intrinsic gains in fitness.
This leads me to the realization that I did not substantiate my off-hand remark about the variants not being intrinsically more fit: At least, not in aggregate. The VOCs were not uniformly successful (natural or unnatural) “experiments.”
So, even though the lesser VOCs have enough a “foothold” to generate a few thousand sequences, thus defining a true clade (as opposed to one-off mutants that land above the trend for mutation distance, but do not form lineages), they are not, in fact, uniformly better at growth vs. the background rates.
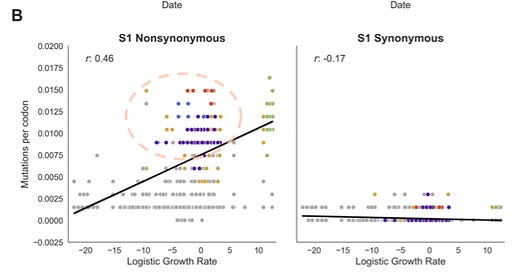
The best visualization here builds off of Trevor Bedford’s presentation of September, 2021, which was followed up in a pre-print this January.1 Bedford’s team is attempting to show that having a lot of nonsynonymous (amino acid-changing) mutation on the S1 head of the spike protein confers a growth advantage.
I would criticize this overly math-y conclusion on many levels, but my purpose here is merely to co-opt their "clade growth" analysis to show that very few of the variant clades were actually more growth-y than pre-VOC clades (using a globally sampled set of sequences from the late 2019 to May 21, 2021).

Gold dots correspond to “other” variants; which although they leap above the trend in terms of mutations vs. ancestral clades (as shown in Neher’s analysis) merely “tie” or even underperform the best of the ancestral clades. And, Gamma and Beta are both middling, if not growth-defective, as well (though, again, this is such a math-y and highly complex way of looking at things that are limits to the meaning of the results). (Regarding “bottom of the barrel” ancestral clades, see footnotes.2)
An even more peculiar observation one can make, drilling all the way down into the last item in the supplemental materials, is that all the clades — pre-VOC or otherwise, exhibit a “fade” effect, with growth slowing as the year goes on.
Wherever they “start” in terms of growth thus likely defines as a real condition the apparent size of their foothold, and as a mathematical artifact their success over the aggregate period in Fig. 1 (Beta, which was never a high-growth variant anyway, is an exception).
I invite the reader to consider whether the following is what would probably be seen if these “infusions” of different genomes of SARS-CoV-2 into the human population have some sort of intrinsic half-life (I actually don’t know the answer to this question, because so much complexity is introduced by Kistler, et al.’s methods here):

By the time Delta has settled in for its autumn, 2021 wave… It is “growing” (however the algorithm defines it) not much more quickly than the dying ancestral clades were in March.
Below is Bedford’s “plain-English” explanation of the method for quantifying growth, followed by the text of the paper. The latter is difficult to parse, to say the least. But based on the former I think all that is being measured is how quickly clades can “gain dots” (resulting in dot-density in a given time and branch).
Therefore (I think), Delta’s “slowing” is not an artifact of it being measured “relative to itself;” but rather of all sub-Delta clades beginning to lose steam at the same time (even though case rates were still cresting).
While this could be consistent with self-competition; it still reinforces the similarity between Delta in autumn and pre-VOCs in spring:
Logistic growth of individual clades was estimated from the time-resolved phylogeny and the estimated frequencies for each strain in the tree. Frequencies were estimated with Augur 12.0.0 [34] using the KDE estimation method that creates a Gaussian distribution for each strain with a mean equal to the strain’s collection date and a variance of 0.05 years. At weekly intervals, the frequencies of each strain at a given date were calculated by summing the corresponding values in their Gaussian distributions and normalizing the values to sum to 1. The frequency of each clade at a given time was the sum of its corresponding strain frequencies at that time.
Logistic growth was calculated for each clade in the phylogeny that was currently circulating at a frequency >0.0001% and <95% and that had at least 50 descendant strains. Each clade’s frequencies for the last six weeks were logit transformed and used as the dependent variable for a linear regression where the independent variable was the corresponding date value for each transformed frequency. The logistic growth of the clade was then annotated as the slope of the linear regression of the logit-transformed frequencies.
If you derived value from this post, please drop a few coins in your fact-barista’s tip jar.
Kistler, KE. Huddleston, J. Bedford, T. “Rapid and parallel adaptive mutations in spike S1 drive clade success in SARS-CoV-2.” biorxiv.org
Kistler, et al. create a mathematical illusion of nonsynonymous S1 mutations being correlated with fitness via a double-outlier effect.
In the first place, Delta is the only extraordinarily high-growth VOC in this period (and we know from the animation, that this will not last anyway). However, they claim that with Delta removed the magic correlation “r” number stays at .41.
The second outlier effect is via their methods for defining (or if you want to say, synthesizing within the data) a “clade” in the Nextstrain sequence batches for the purposes of the growth analysis:
Logistic growth was calculated for each clade in the phylogeny that was currently circulating at a frequency >0.0001% and <95% and that had at least 50 descendant strains [I believe “strains” means sequences here, “strain” just being what the value is called within the tool].
Theoretically, these cutoffs should be indifferent to biases or artifacts. In fact, what happens is that the cutoff operates like a resolution filter. With bigger data (amount of clades), it will offer a higher resolution, and capture more close-to-baseline clades. With fewer clades, it will offer a lower resolution, and not capture close-to-baseline clades.
In effect, all that is happening here is that there are fewer high-S1-mutation clades (VOC or otherwise), because they do not appear frequently, and a lot more 1-S1-mutation clades, because they will branch off from the root S1 sequence all the time, and so the cutoff is capturing the rare borderline cases in the 1-S1-mutation clades and no rare cases in the high mutation clades.
This was a bit of a mind-trip for me to figure out, as I knew there had to be some kind of bias for “more successful clades” in the higher mutation ranks, but I couldn’t figure out how the bias didn’t “confirm the result.” It is (to restate) because rare near-cutoff-success, low-mutation clades are being captured just over the cutoff by sheer volume of all low-mutation clades, and then incorrectly being weighted as “one” vs. the higher success low-mutation clades in calculating the correlation.






One explanation for clades that appear and then fade out is that they were intentionally "seeded" within a population. I'm assuming this is where you are headed with this logic, and I almost suggested the same thing in my comment on your last post.
I'm curious, though, whether this phenomenon (many short-lived, local/regional evolutionary clades which fail to exhibit greater fitness on a global scale) is the exception or the rule in viral evolution - or even in evolutionary contexts more broadly.
The vast majority of mutations will be deleterious and will not be passed on from the host where they arise. A select few will confer greater fitness and will eventually gain dominance across the global viral population.
What lies in between these outcomes, though?
1. Mutations that confer a selective advantage *only within the host where they arise*. Perhaps they are better at infecting cells or making more copies but terrible at being transmitted. Presumably this explains many of the "one-off" sequences that are detected.
2. Mutations that confer a selective advantage within a particular spatiotemporal context, but not globally/permanently. It's quite possible to imagine, for example, a strain that could outcompete existing strains (and cause a wave of infection) in Manaus but not in New York City, or vice versa - in the context of differing innate/adaptive immunity in the populations (from past exposure to different pathogens/antigens) and different environmental and social conditions influencing transmission. It's also possible to imagine a strain that has an advantage in northern hemisphere winter but not in northern hemisphere spring/summer.
One of the main challenges here is that we have more detailed sequence data for SARS-CoV2 across space and time than for any other virus or organism ever, and so it's not always easy to find parallel comparisons to assess what is simply revealing the micro-, meso-, and macro-dynamics of natural selection and what is evidence of artificial intervention.
https://palexander.substack.com/p/ba5-covid-sub-variant-being-replaced?pos=1
------
"A non-sterilizing vaccine WILL cause variants to emerge, as this is doing!"
So this is Paul channeling GVB, and Paul and GVB are quasi-homies or at least simpatico on this theme.
-----
Brian, I will confess to diligently trying to read through the first post in the series. I was doing pretty good through the Tony Hawk part and stuff. My head exploded, which is not unusual.
----
How far apart are we here on the Expert Scale?
Polar opposites?