Should variants actually happen? Pt. 1
Reexamining the giant assumption that was plugged into our heads in advance.
Decades of popular mongering of the current human understanding of evolution have created the widespread impression that microbes and humans are more-or-less always in an “evolutionary arms race.” Human fires weapon at microbe; microbe evolves new defense against weapon; like the Borg, in the 90’s Star Trek show.
In fact, there are myriad counter-examples to this assumption of evolutionary dynamism, and when it comes to the textbook “example” for viruses — flu — matters aren’t actually as dynamic as they seem. These data points, however, are typically just forgotten about since they contradict the biological narrative in vogue.
Also forgotten, is both that SARS-CoV-2 barely mutated at all in 2020, and that the heralded “variants of concern” that arrived from nowhere at the end of the year were not immune-evasive.
This post is therefore an attempted, definitely non-copyright-infringing excursion in viral evolutionary myth- “boffing” (“boff” is in the thesaurus entry for “bust”). I think that boffing these myths is necessary before considering the question of whether any of the VOCs had a natural origin directly. So let’s do it!

Part 1 (here) will address the first myth.
In Part 2: A bonus myth, regarding the “coronavirus pandemic” of 1889, and general evidence of coronavirus genetic stability.
In Part 3: Genetic stability in SARS-CoV-2, and the problem with its evolutionary trend.
No, But Seriously, Set Down Expectations about Evolution, Swarms, etc.
Really — set them down! Otherwise I fear that even simple statements are prone to misinterpretation, because the reader will imagine that I must be confirming whatever it is that they think, in advance, I have come to reveal. In fact, I am probably not confirming what it is you think in advance I have come to reveal.
And so I urge that the reader prepare to take all the following at face value. The three myths above really are myths.
A lot of viruses, including RNA viruses, don’t mutate that much. The ones that do are exceptions that prove the rule.
This fact — that SARS-CoV-2 would only be dynamic if it were exceptional — is, for a preview of the coming post regarding whether the SARS-CoV-2 variants of concern are natural, the very reason for any hypothetical human manipulation of the virus after the initial “outbreak.” Consider the following sentence from an early review of the virus’s evolutionary behavior in the first 2020 wave; think of the implications:
Although we cannot predict whether adaptive selection will be seen in SARS-CoV-2 in the future, the key finding is that SARS-CoV-2 viruses that are currently circulating constitute a homogeneous viral population. Viral diversity has challenged vaccine development efforts for other viruses such as HIV-1, influenza, or Dengue, but these viruses each constitute a more diverse population than SARS-CoV-2 viruses. We can therefore be cautiously optimistic that viral diversity should not be an obstacle for the development of a broadly protective SARS-CoV-2 vaccine, and that vaccines in current development should elicit responses that are reactive against currently circulating variants of SARS-CoV-2.1
But-but-but-but vanden Bossche said!
I know! He told you that the vaccines would create an escape variant, so it should make perfect sense that “escape variants” appeared after the deployment of the vaccines. And this accorded exactly with the drip-feed of pop science narratives about human-microbe evolutionary arms races that has been fed to humanity for decades.
But dispelling illusions about some fatalist, guaranteed failure of the vaccines because evolution is precisely the reason for this post. Vaccines fail because they are a stupid, primitive attempt to improve a system that is better left not-interfered-with. The case for any, ever having failed “because evolution” is in fact weak. Hence why this post will also discuss why influenza does not even belong in the list of exceptions.
RE swarms
This post will to some degree refute the argument made by JJ Couey for multiple or perpetual-ish releases.2 Instead of emphasizing RNA instability, I will be emphasizing stability. The point, however, is still to form an argument for multiple releases.
The reader may think of this as an alternate model of the “Gain of Purity” theory. My goal, in other words, is to make the theory more robust by ensuring that there are bets placed on both sides of the RNA virus fidelity question, rather than leaving the chips all on red.
Still, it bears repeating, that the reader should set down any expectations of this being a post in which swarms are secretly very meaningful; they aren’t that important here.
i. RNA Fidelity via Stabilizing Selection
Arms Race Counterexamples: Microbes are not Borg
First, some baseline-calibration. Examples of microbes “evolving” in response to human efforts at sterilizing them are not, in fact, common. None may truly even exist.
In bacteria.
Antibiotic resistance is not the product of bacteria mutating or evolving in a sense of true progress or novelty. Instead, resistance genes exist in the bacterial gene pool in advance, and are likely ancient (which makes sense, since we poach antibiotic molecules from other microbes that have been competing with bacteria for eons). The genes are modular and can move horizontally in bacteria via plasmids or viruses (phages). From a 2020 review: 3
The most clinically significant [antimicrobial resistance genes] are usually located on different mobile genetic elements (MGEs) that can move intracellularly (between the bacterial chromosome and plasmids) or intercellularly (within the same species or between different species or genera). […]
The phenomenon of antimicrobial resistance (AMR) is not new as ARGs have evolved over millions of years (Shlaes et al., 1997). However, AMR is amplified in the presence of the selective pressure exerted by antibiotics
Selection pressure can exert a bottleneck effect on pre-existing, million-years-old resistance genes that move horizontally in bacterial genomes; it cannot create them out of nothing.
The modern trope of fretting about an antibiotic-resistance apocalypse, where all bacteria on Earth are turned into permanent super-bugs, is thus totally overblown. At worse, there may come a time when antibiotics have to be taken out of circulation for a few years until the global bacterial population returns to equilibrium in terms of plasmid / phage gene distribution; the waters should calm themselves if we simply stop stirring for a while. But even this day may never come.
On the other hand, hospitals, with their constant sterilization, will probably remain hotbeds of resistance gene-modded bacteria no matter what medication choices humans make outside.
So, total bonus myth-boff, there. (But antibiotics may still be a net neutral or net negative for humans due to environmental contamination, disruption of the microbiome, etc.)
In RNA viruses.
Polio virus, unlike Measles, does not require passage through the bloodstream for successful transmission. Neither the oral nor injected polio vaccine are confirmed to be sterilizing -- they are, rather, true “leaky” vaccines; and yet decades of use of both have failed to create an “escape” variant. (Regarding the vaccine-derived polio viruses, these are viable viruses that are selected from a bottleneck even during OPV administration, and are only one mutation away from the expected vaccine genomes.4 So this is also not an example of dynamism in polio.)
The question of how the injected vaccine can even lead to the apparent local eradication of the virus if it is not sterilizing, I will leave aside for now. My point is simply that this RNA virus has been given a “leaky” obstacle and yet has not evolved its way around it, despite decades. The likely reason is that polio is constrained by a force called stabilizing selection.
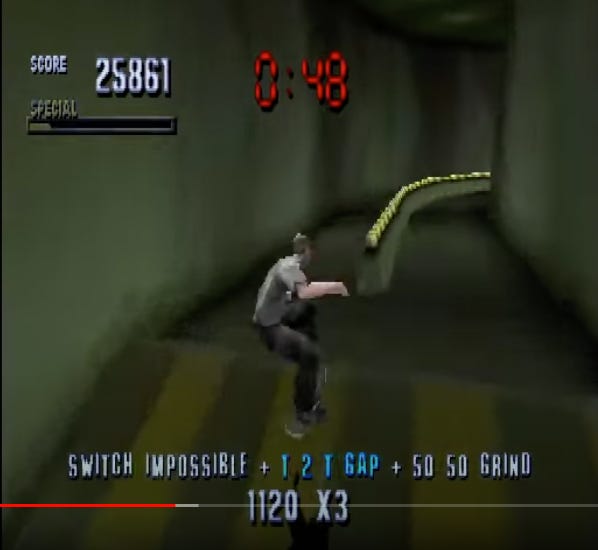
Stabilizing Selection in RNA viruses: The Tony Honk Model
In the definitely not copyright-infringing classic video game Tony Honk’s Pro Skater, the player is tasked with wandering around urban settings and maximizing the amount of cool stunts that can be performed in a given time. Stunts are performed by selecting between the four buttons on your Playstation controller. Just like genes can code using four nucleotides. So, there is a limited spectrum of possibilities.
For any given element in the map, there is a maximum possible score that can be achieved corresponding to an optimum move order during interaction with the element.
In the example Tony Honk Pro Skater (THPS) screen-grab above, if we imagine that 1120 x 3 is the maximum possible score for interacting with and jumping between these two rails, then “Switch Impossible + T 2 T Gap + 20 20 Grind” are the optimum move set (or within a group of optimum moves). No other move set will outperform them.
This means that two separate players wanting to achieve 1120 x 3 here will have to do it the same way. Supposing that two players, who happen to be parent and child, do do so — then where is the “information” for the optimal move stored? In their genes? No, it is stored in the game element itself.
The THPS model also prevails for viruses. The most successful at “jumping” through any given course — replication cycle — will always be those that perform the optimal move set; which will always be those with roughly the same genes; because the genes are the “moves.” The virus’s genes are thus a mirror of the host biology; just as the THPS player’s moves are a mirror of whatever element is being interacted with. Another analogy might be sonic resonance; if RNA viruses are imagined as being akin to a harmonic sound arising from our shared cellular receptors, their tones and wavelengths are determined by the substrate, i.e. us. Anything that is noise cannot resonate, and so our viruses remain “pure” throughout time.
This force is known as stabilizing or purifying selection (there is a semantic difference between the two, but in the THPS model there is no reason to make a distinction). The force works in concert with the viral “swarm” to produce sustained genetic stability, rather than stability being rendered forbidden by it. Even if 99,999 viral progeny are lost (or fated to fade away) due to inheriting suboptimal mutant THPS moves; the perfect copy, or the reversion (recovery mutation) to the perfect model, survives in the long term. From Edward C Holmes’s Evolution and Emergence of RNA Viruses:5
As stressed a number of times already, the most likely explanation is that RNA viruses possess very large population sizes for much of their life cycle (although this clearly does not apply to latent viruses). Hence, although deleterious mutations are produced in very large numbers, so are the viral progeny generated during every replication cycle. Large populations also mean that there will be sufficiently frequent reversal and compensatory mutations to off-set the accumulation of deleterious mutations. In short, RNA viruses are able to produce greater numbers of offspring than are removed by purifying selection, and it is this massive reproductive rate that is the key to their evolutionary survival.
Where this force becomes compromised, of course, is in the context of adaptive immune recognition. If the former optimum set of moves is arbitrarily rendered unfit, then viruses, just like a THPS player, will have to find whatever is the alternate set of moves that leads to the highest possible score, even if it is not 1120 x 3. Here the immune system acts like a bored audience that tires of THPS perfection. A move-set producing a 1000 x 3 score will become selected purely for novelty, despite being suboptimal in a perfect system.
Stabilizing Selection Exemplified: Influenza A Virus
And so it turns out that the organism most frequently cited as a precedent for “endless immune escape” also exemplifies the default to stability in the RNA virus model.
Stasis in earliest H1N1 isolates.
A fascinating example is “PR8,” the second-ever recovery or “isolation” of a human flu, transferred first from human to ferret in 1934, and then passaged to mice, and operating since then as a mice-adopted antigenic mannequin of influenza A as it existed in the mid-30s.
Because ferret, then mice passage was required to acclimate PR8 to its new host, we can presume some early genetic remodeling, i.e. “gain of function.” But what is remarkable is that no matter how many times the virus has been passaged afterward, it remains self-referential in terms of physiochemical (antigenic) design: A mouse given one lab’s copy of PR8 will make antibodies that can defend another mouse against another lab’s copy. Likewise, PR8 remained a model for human flu memory responses for people alive in the 1930s for as long as anyone bothered to check.
This same stability prevails in the classic “swine flu” spike protein (HA) gene, which remained fixed as it naturally rebounded from generation to generation in American hog farms for almost a century after 1918. Because successive generations of hogs did not have immune memory, the virus’s spike protein did not evolve. For more on all of the above, see my reconstruction of the early history of flu research:
Only the introduction of anti-PR8 antibodies can induce the PR8 flu virus to deviate from its stable optimum. Even more impressive, PR8 has been shown to regress back to the optimum if, first, confronted with an immune response that increases receptor binding, and then re-introduced to non-immune mice:6
Optimizing viral fitness requires balancing host cell receptor binding of input virus with release of progeny virus. Strikingly, passaging in vivo [vaccinated-mice-passage] selected mutants in naïve [unvaccinated] mice selected [2nd generation] mutants with reduced cellular receptor binding avidity. Mutations selected by naïve mouse passage decreased polyclonal HAI antibody escape.
The experiment above illuminates the fact that the “optimum” for influenza is not always to bind to sialic acid as strongly as possible, because this may impede release from previously infected cells — the springiness of the “jump” in the THPS model. Immune responses force influenza to favor a tighter-than-optimal snap to the rail in order to stick the landing.
Stasis in wild influenza A.
Decades of genetic analysis have found that most flu genes are molecularly inert in any given host; and that the spike protein (HA) gene is only dynamic in humans, due, it is safe to presume, to our persistent immune responses which do not feature in the avian, equine, or swine flu ecosystems.
Here I could quote extensively from the classic sermon lead-authored by the Pope of flu, Robert Webster. One selection will have to do:7
Homologous sets of genes from host-specific virus strains also evolve at different rates. Recent evolutionary studies of influenza virus NP [nucleoprotein, the flu RNA-winding-and-packing-up protein] genes have shown that avian virus genes are evolving more slowly than those in human viruses and that avian virus proteins are highly conserved, showing no net evolution over the past 60 years. Within the Old World avain virus NP gene lineage there is no accumulation of amino acid or coding changes; all accumulating nucleotide changes are silent. Classic swin virus NP genes are evolving similarly to human virus NP genes, but at the protein (amino acid) level the swin virus NPs are more conserved. Genes of H2N5 equine 2 viruses are evolving more slowly than human or swine viruses.
Different flu genes evolve at different rates, depending partially on the presence of host immune pressure (in the case of humans). But even in humans, there are examples of near-perfect stasis, such as for the “matrix” protein. When the genes of 1930’s and later recovery-passage-preserved models of flu matrix protein are compared, there are almost no differences in the amino acid “moves” called for by the M1 gene, despite millions of intervening replication events:

The degree to which stabilizing selection also prevails for “silent” gene mutations — where a different THPS button is pressed but the player makes the same move — will be discussed further in the context of SARS-CoV-2.
Stasis and flu recycling.
The reality illuminated by this evidence, when considered in totality, is that flu only evolves in the face of immune pressure; and that there is a finite amount of possible alternate “moves” before flu becomes patently unfit. Eventually, once any model of flu has been in human circulation for a long enough time, it runs out of moves that even get it to the next rail. This is why flus vanish from circulation:
And so when “novel” flu spike protein genes re-enter the human circulation, they are not in fact novel at all. They are simply recycling in from the avian reservoir of genetic flu virus stasis. They are akin to “back-ups” of the original version of the spike protein that retain the optimum move-set that was lost in the human flu ecosystem; again, since water fowl do not mount immune pressure on flu, the spike protein is also preserved in its “pure” form by stabilizing selection.
As there is evidence that the 20th century’s H1, H2, and H3 spike protein variations were all featured in the flu outbreaks before 1918, it remains to be seen if any of the other spike protein models are even compatible for human-to-human transmission (without an implausible degree of genetic remodeling). Instead, the modern era has simply consisted of a slow rotation between those three models. A definitely not-weird way to visualize the interplay between the avian and human flu ecosystems, is to imagine the avian flu genes as cosmic, timeless jellybeans that occasionally condense and fall onto the landscape of human life, only to quickly evaporate:

In sum, the hyping of flu’s endless genetic dynamism is just that: Hype, without much substance. While antigenic “drift” might be impressive in a lab setting, and certainly seems plausibly related to seasonal flu resurgence, it is a short-term solution to immune memory that eventually must succumb to the inevitable. To remark briefly on vaccine evasion, the evidence is in fact mixed whether drift has any relationship to vaccine failure, as opposed to a “no pain, no gain” model where the vaccinated only get durable immunity when they stop delaying inevitable infection.8
Regardless, drift in human flus is a temporary scheme exhausting a finite spectrum of possible alternate spike protein builds, with the current model of flu spike protein eventually no longer able to spread in the human ecosystem — which is why every spike protein cycle of flu before the current two died out (no comment, for the moment, on whether the current two are being accidentally sustained by constant human fussiness over isolating and testing them).
Meanwhile, “antigenic shift” — the introduction of alternate spike protein models from the avian reservoir — is no more a marker of novelty than whatever decade’s pop song sampling “Genius of Love” is. It’s just the same old genes, appearing novel to new generations, forever. What is often marketed as the most radical form of genetic change in flu is in fact a marker of stasis.
Summary: Are there any truly dynamic respiratory viruses?
If even flu turns out to be genetically stable, it begs the question of whether it is even possible for respiratory or enteric viruses — the staples of “childhood infections” — to persist in the “immune-evasive” model. Recall that the other two examples of antigenically dynamic viruses, HIV and dengue, are essentially unicorns. They spread by unique means; they occupy and interact with their hosts in a manner more akin to a parasite lifecycle than a childhood virus. (This might also obtain for Ebola and other modern monsters-of-the-week, but I still have yet to look into those topics.)
Meanwhile, polio, measles, and other RNA viruses seem indifferent to the evolutionary challenge posed by modern vaccination. If they cannot have their established, optimum designs, they would just as soon go extinct. For these viruses, in other words, there exists only one way to play their chosen course in the skate-park of human biology, and we have made it impossible. Naturally, we should expect that these viruses might still achieve immune escape eventually, but it will remain true that the process took decades.
Whereas, for flu, it is first true that the virus can only survive thanks to the avian reservoir, and second true that nothing is really novel or surprising about crossover of the three known spike protein (HA) genes into humans. It has probably been happening forever, because human-compatible viral genes in the animal reservoir will cross over easily. (Which means, by the way, that flu is not evidence that we should expect random new viruses to “emerge” every time we turn on the news. We should be suspicious of novel viruses. More on this in the coming segments.)
Given that everything about flu is sui generis, it cannot be used as a model for other respiratory viruses without an abundance of caution. It is, however, a valuable reference simply because it has been studied and genetically sampled more extensively through time than any other human virus: And in that manner, it is proof of the potential, if not propensity, for RNA virus stability.
And, although it has not also been around for decades, so is SARS-CoV-2.
To be continued in Part 2…
If you derived value from this post, please drop a few coins in your fact-barista’s tip jar.
Dearlove, B. et al. “A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants.” Proc Natl Acad Sci USA. 2020 Sep 22; 117(38): 23652–23662.
See Crawford, Mathew. “SARS-CoV-2 Origins, Infectious Clones, Biowarfare, and Robert Malone.” (2022, November 7.) Rounding the Earth.
See Vrancianu, CO. Popa, LI. Bleotu, C. Chifiriuc, MC. “Targeting Plasmids to Limit Acquisition and Transmission of Antimicrobial Resistance.” Front Microbiol. 2020; 11: 761.
Cann, AJ. et al. “Reversion to neurovirulence of the live-attenuated Sabin type 3 oral poliovirus vaccine.” Nucleic Acids Res. 1984 Oct 25; 12(20): 7787–7792.
Edward C. Holmes. The Evolution and Emergence of RNA Viruses (Oxford Series in Ecology and Evolution) (p. 54). Kindle Edition.
Hensley, SE. et al. (2009.) “Hemagglutinin Receptor Binding Avidity Drives Influenza A Virus Antigenic Drift.” Science. 2009 Oct 30; 326(5953): 734–736.
Note that the regression is via a secondary mutation which returns binding affinity to baseline, rather than a reversion of the first mutation. This suggests that the pure PR8 model was lost from the “swarm.” This may mean that the regression mutant is still sub-optimal due to having accidentally chosen a node that further widens the fitness valley separating it from the original version. In a natural or wild flu ecosystem, it would likely still die out, leaving the pure version. This is again why I don’t think it is too productive to treat stabilizing and purifying selection as separate forces; both are attempting the same end and the one that will obtain simply depends on the dynamics of the system as a whole (does a regression mutant have to compete with the original allele or not).
Webster, RG. Bean, WJ. Gorman, OT. Chambers, TM. Kawaoka, Y. (1992.) “Evolution and Ecology of Influenza A Viruses.” Microbiol Rev. 1992 Mar;56(1):152-79.
The irony here is that no one has done more to promulgate the 21st-century paranoia of a future avian flu crossover than Webster, even though he is responsible for the vast repository of genetic evidence demonstrating that avian flu is in genetic stasis! I think that 90s Star Trek got to him, too.
Discussed tangentially in “OAS Lit. Review/Timeline, Pt. 2.”










Can we vote for a show w you & JJ slow walking this w graphics maybe for folks in the bleachers?
Your thinking in these matters continues to parallel mine. My background, however, is in computer science (BSCS plus 50+ years in the field; couldn't find a use for an advanced degree). There is a surprising degree of overlap. I became interested in the mechanisms of protein synthesis about 10 years ago, and it may have saved my life in 2021, if you see what I mean.
I can't pause for a lengthy reply today, but I'll challenge you with one question: Could your view of viral behavior offer a clue that they may have been _designed_ this way? It's surprising what can pop out when you revise your assumptions.